Hanna Vuorenpää1, Roope Ohlsbom1, Kaisa Tornberg1, Elina Kalke1, Jonathan Sheard², Pasi Kallio¹, Susanna Miettinen¹
1Tampere University, Faculty of Medicine and Health Technology, Tampere, Finland
² UPM Biomedicals, Helsinki, Finland
GRAPHICAL ABSTRACT
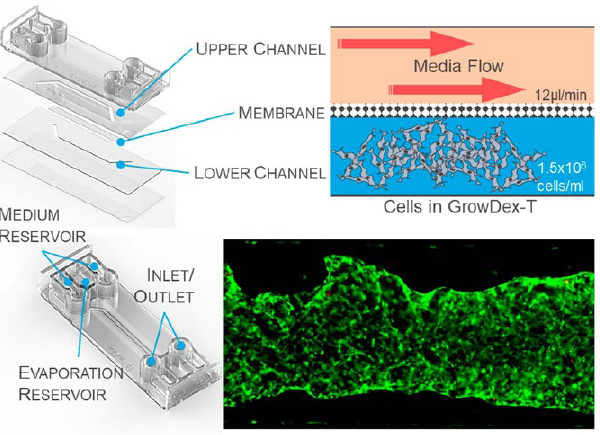
Figure 1. The BE Doubleflow microfluidic chip from BEOnChip has two channels which allow microfluidic flow, and they are separated by a porous membrane. BMSCs (1.5x10^6 cells/ml) were embedded within GrowDex-T and injected into the lower chamber. Media was loaded into and flowed through the upper chamber at 12 µl/min.
INTRODUCTION
Conventional two-dimensional in vitro setups often face challenges in recreating the required complexity of the cell’s microenvironment [1, 2]. Cell-laden hydrogels offer much more representative models of in vivo tissues compared with two-dimensional cell cultures [3], bridging the gap between in vitro and in vivo culture conditions. Today, the most widely used hydrogels are animal-derived which exhibit lot-to-lot variability, poor definition and understanding of composition and temperature sensitive crosslinking [3, 4]. Additionally, these animal derived hydrogels fail to represent the human system sufficiently [5].
GrowDex-T is anionic nanofibrillar cellulose hydrogel derived from birch. It is temperature stable, requires no cross-linking or gelation steps, it has reproducible lots, and is compatible with many cell types [6]. Thus, GrowDex-T could offer a convenient, animal-free option for recreating the complexity of the cell’s microenvironment. In this collaborative study, we wanted to test the compatibility and suitability of GrowDex-T as a 3D microenvironment for culturing mesenchymal stem cells in a commercial organ-on-chip (OOC) device under active flow. Alongside GrowDex-T, fibrin and collagen type I were similarly tested.
The BE-Doubleflow (Figure 1) chip from BEOn Chip (Spain) was chosen due to its unique dual chamber design, allowing for the cell-laden hydrogels to be loaded into the lower chamber, whilst media flowed through the upper chamber. Initial cell culture tests were performed, to identify an optimum flow rate, seeding density, cell type and culture time. Here we present results of a 7day culture of BMSCs seeded at 1.5x106 cells/ml with a media flow rate of 12 µl/min, embedded in GrowDex-T, fibrin and collagen type I.
The content presented in this application note is a continuation from Application Note 11, ‘GrowDex-T in a commercial organ-on-chip under active flow [7], therefore for repeated methods please refer to the previous application note.
MATERIALS
Cells, Organ-on-chip device and Hydrogels
- BMSCs (Primary bone marrow aspirate, Tampere University Ethics code: R15174)
- Be-Doubleflow standard (BEOnChip)
- GrowDex®-T, 1.0% (Cat. No. 200 103 005, UPM Biomedicals, Finland)
- Fibrinogen from human plasma (Cat. No. F3879, Sigma-Aldrich)
- Thrombin from human plasma (Cat. No. T4394, Sigma-Aldrich)
- Aprotinin, serine protease inhibitor (Cat. No. ab146286, Abcam)
- Collagen I, rat tail, Gibco™ (Cat. No. A1048301, ThermoFisher)
Flow platform, connectors, tubing, and accessories
- Ismatec IP peristaltic pump, 12 channels
- Tygon® Tubing 0.5mm ID x 1.5mm OD (PVC) (Cat. No. T4102, Qosina)
- HPLC tubing, Upchurch Scientific®, 1/16” OD x 0.030" ID x 100ft (Cat. No. 554-2980, VWR)
- BEOnChip peristaltic/syringe pump connectors, 1/4-28”, barb 1/16”
- BEOnChip inlet/outlet plug, 1/4-28”
- Ismatec PharMed® BPT 0.19mm ID orange/red, 2-stop (Cat. No. WZ-95723-10, Cole-Parmer)
- Silicon tubing 1mm ID x 3mm OD (Cat. No. 228-0701, VWR)
Other reagents and laboratory accessories
- Complete media (CM):
- αMEM, no nucleosides, Gibco™ (Cat. No. 22561021, ThermoFisher)
- Human serum, used at 5% (Cat. No. S-HU-EU-011, Serana)
- Penicillin (10,000U/ml) & Streptomycin (10mg/ml) (Cat. No. DE17-602F,
Lonza))
- Human FGF-2 (Cat. No. 130-093-840, Miltenyi Biotec)
- TrypLE™ Select Enzyme (1X), no phenol red, Gibco™ (Cat. No. 12563011, ThermoFisher)
- 200 µl graduated TipOne® Filter Tip (Cat. no. S1120-8710, Starlab)
- 1,000 µl XL graduated TipOne® Filter Tip (Cat. No. S1122-1730, Starlab)
- TC20 automated cell counter (Bio-Rad Laboratories)
- 1 x DPBS without phenol red (Cat. No. 17-512F, Lonza)
Staining reagents
- LIVE/DEAD™ Viability/Cytotoxicity Kit, for mammalian cells, Invitrogen (Cat. No. L3224, ThermoFisher)
- Phalloidin–Tetramethylrhodamine B isothiocyanate (TRICT) (Cat. No. P1951, Sigma-Aldrich)
- 4′,6-diamidino-2-phenylindole (DAPI) (Cat. No. Sigma-Aldrich D9542)
- Paraformaldehyde (PFA) (Cat. No 15713, Electron Microscopy Sciences)
- Bovine serum albumin (BSA) (Cat. No. A7906, Sigma-Aldrich)
- Triton X-100 (Cat. No. T8787, Sigma-Aldrich)
Microscope and image processing
- Olympus IX51 Fluorescence Microscope
- Olympus cellSens Dimension Imaging Software
- ZEISS Primovert Inverted Microscope
- ZEISS Axiocam ERc 5s
- Fiji-ImageJ-64bit, v1.52p
METHODS
Overview: BMSCs were embedded in 0.5% GrowDex-T, 2.5 mg/ml fibrin, or 2 mg/ml collagen type I at a density of 1.5 x 106 cells/ml cultured for 7 days (long-term) in BE-Doubleflow chips. Aprotinin was added to the fibrin hydrogel (12.5 µg/ml) to prevent gel degradation.
First, cell-laden hydrogel was loaded into the bottom channel of BE-Doubleflow chip, then media was added to the upper chamber for 24hrs, then pumped through as active flow by peristaltic pump for 7 days. Parallel static controls were established as well. Instead of connecting to the peristaltic flow, the static control chips were placed into humidified chambers and medium was changed manually on a daily basis.
Hydrogel integrity and cell morphology were assessed daily using brightfield microscopy. The chips were not disconnected from the peristaltic flow at any point when the experiments were ongoing. At the end of both experiments cell viability was assessed with live/dead assay. At the end of the long-term culture, phalloidin staining was performed to assess cell morphology and alignment. Qualitive analysis of the results was conducted using fluorescent microscope.
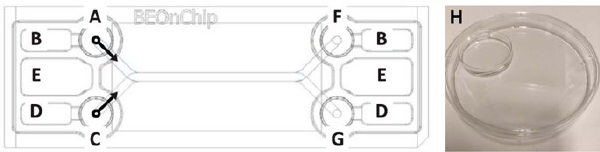
Figure 2. Top view of BE-Doubleflow chip: A) Upper channel inlet and inlet reservoir, B) upper channel medium overflow reservoir, C) bottom channel inlet and inlet reservoir, D) bottom channel medium overflow reservoir, E) evaporation reservoir, F) upper channel outlet and outlet reservoir, G) bottom cannel outlet and outlet reservoir. Modified from BE-Doubleflow technical characteristics data sheet, H) Humidity chamber made with petri dishes for BE-Doubleflow chips.
Loading BE-Doubleflow with GrowDex-T:
- According to the manufacturer’s instructions, prior to addition of the hydrogel or cells, incubate the empty chips for at least 2 days at 37°C and 5% CO2 in humidity chambers i.e., large petri dishes with smaller petri dishes inside filled with H₂O (Figure 2 H).
- Add 400 µl of 1 x DPBS into the evaporation reservoirs of chips (Figure 2 E).
- Prepare and count BMSCs into a cell suspension with a minimum concentration of 4.0 x 106 cells.
- Prepare 2 ml of GrowDex-T (0.5%) embedded with BMSCs (1.5 x 106 cells/ml) according to the GrowDex-T “Instructions for use” protocol [6]. This can be done by following the protocol detailed below.
- Add 800 µl of CM at room temperature (RT) into a 2.0 ml Eppendorf tube.
- Weigh 1000 mg of GrowDex-T (stock 1.0% at RT) into the Eppendorf tube on a scale.
- Mix the GrowDex-T and CM thoroughly using a 1000 µl pipette with low retention pipette tips until the solution appears homogeneous.
- Take a required amount of the BMSC suspension with 3.0 x 106 cells; centrifuge (200 g, 5 min) and resuspend in 200 µl of CM to.
- Add 200 µl of the prepared BMSC suspension to the GrowDex-T dilution to obtain a final concentration of 0.5% GrowDex-T dilution with a cell density of 1.5 x 106 cells/ml. Mix carefully using a 1000 µl pipette with low retention pipette tip to ensure homogenous distribution of the cells in GrowDex-T.
- Add 100 µl of cell-laden GrowDex-T into the bottom channel from one side using a 1000 µl pipette (Figure 2 C).
- Add 50 µl of CM (37°C) into the bottom channel inlets to prevent drying (Figure 2 C and G).
- Incubate the chips at 37°C and 5% CO₂ in humidity chambers for 30 minutes before adding the media
Loading BE-Doubleflow with cell-laden fibrin
Fibrin hydrogel (2.5mg/ml) embedded with BMSCs (1.5 x 106 cells/ml) was prepared as previously described [8] with a few modifications. This can be done by following the protocol detailed below.
1. Prepare 2 UI/ml thrombin solution in CM and 5 mg/ml fibrinogen solution in DPBS.
2. Add aprotinin to the 5 mg/ml fibrinogen solution so that the aprotinin concentration is 50 µg/ml to prevent degradation of the gel
3. Take a required amount of the BMSC suspension; centrifuge (200g, 5 min) it and resuspend the BMSCs in 2U/ml thrombin at a concentration of 3 x 106 cells/ml.
4. Divide the cell-thrombin mixture into 75 µl aliquots.
5. Pipet 75 µl of fibrinogen-aprotinin mixture into an aliquot of BMSC-thrombin mixture. Mix quickly and continue onto loading the gel into the chip (Note, resulting fibrin solution should be loaded to the chips immediately as gelation occurs quickly after fibrinogen and thrombin are combined)
6. Add 100 µl of cell-laden fibrin into the bottom channel from one side using a 1000 µl pipette (Figure 2 C).
7. Add 50 µl of CM into the bottom channel inlet reservoirs to prevent drying (Figure 2 C and G).
8. Incubate the chips at RT in humidity chambers for 20 minutes before adding the media.
9. Add aprotinin to a concentration of 12.5 µg/ml to the media to be used with fibrin hydrogel.
Loading BE-Doubleflow with cell-laden collagen
- Prepare 2 mg/ml collagen solution following manufacturer’s instructions [9]. This can be done by following the protocol detailed below. All the work should be done on ice and cold pipet tips should be used
- Mix 150 µl of 10X DPBS with 25 µl of 1M NaOH and 325 µl of dH2 O in 2.0 ml Eppendorf tube.
- Add 1 ml of 3 mg/ml collagen into the Eppendorf tube and mix the solution thoroughly by pipetting gently up and down.
- Measure pH of the resulting mixture using pH paper. The pH value should be between 6.5 and 7.5 (optimally 7.0).
- Take a required amount of the BMSC suspension prepared in section 2.2.1; centrifuge it (200 g, 5 min) and resuspend the BMSCs in the 2 mg/ml collagen solution at a concentration of 1.5 x 106 cells/ml.
- Add 100 µl of cell-laden collagen I into the bottom channel from one side using a 1000 µl pipette (Figure 2 C).
- Add 50 µl of CM (37°C) into the bottom channel inlets to prevent drying (Figure 2 C and G).
- Incubate the chips at 37°C and 5% CO₂ in humidity chambers (Figure 2) for 30 minutes before adding the media.
Addition of media into the chips
- After the gelation/incubation period, fill the top media channel by adding 100 µl of CM (37°C) into the upper channel using a 1000 µl pipette (Figure 2 A) until the media moves through the channel outlet (Figure 2 G).
- To fill the inlet and outlet reservoirs, add 300 µl of CM (37°C), 100 µl at a time, into the upper channel inlet reservoir (Figure 2 A). The chip should be tilted between the medium additions to allow it to flow through to the outlet side of the chip (Figure 2 F). Media may overflow into the medium overflow reservoirs.
- Image the chips using a brightfield microscope.
- After medium additions, BE-Doubleflow chips should be incubated at 37°C and 5% CO₂ in humidity chambers (Figure 2) for 24 hours before starting the flow.
- After the 24 h incubation image the cells using a brightfield microscope.
Connecting the chips to the peristaltic pump flow
- The flow set-up should be constructed as shown in Figure 5 B. The pumps are placed outside the incubator. The CM, media tubes and connection sites for chip inlets and outlets are placed inside the incubator (37°C and 5% CO2). Each cell culture condition should have their own media tube (with CM at 37°C). The CM to be used with fibrin hydrogels should be supplemented with 12.5 µg/ml aprotinin.
- Sterilize pump tubing by pumping through 70% ethanol and rinse by pumping through dH2O.
- Prime pump tubing with CM (37°C). Set the pump to minimum rate (0.11 rpm) while fixing the chips, connectors, and plugs to avoid trapping air into the flow system.
- Screw the plugs onto the medium-filled bottom channel inlet and outlet reservoirs (Figure 2 C and G).
- Screw the connectors onto the medium-filled upper channel inlet and outlet reservoirs (Figure 2 A and F).
- Manually fill the upper channel with medium by pushing through CM from the inlet connector to the outlet connector (Figure 2 A and F) using a 100 µl pipette with piece of silicon tubing placed on the pipet tip. This will prevent bubble formation in the medium channel.
- Aspirate overflowing medium from the medium reservoirs (Figure 2 B and D).
- Connect primed pump tubing onto the top channel inlet and outlet reservoirs via connectors (Figure 2 A and F).
- Set the pump to a working speed of 2.01 or 5.01 rpm which correspond to flow rates of 12 and 30 µl/min, respectively.
- After starting the flow, image the cells using a brightfield microscope.
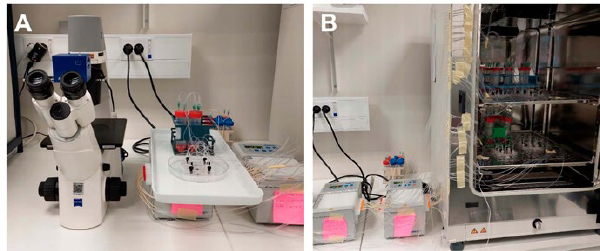
Figure 3. BE-Doubleflow chips and medium tubes were connected to Ismatec IP peristaltic pumps (B) and during the culture period, the BE-Doubleflow chips were imaged daily and moved to the microscope (A) in units of two media tubes and three or four chips.
Cell culture and endpoint analyses
- Image the chips daily using a brightfield microscope. Keep the chips connected to the flow when moving them to the microscope. This can be done by moving the chips and media tubes as units of two tubes and three or four chips (Figure 3 A and B).
- Once the experiment is over, disconnect the chips from the flow and perform the necessary staining, such as Live/Dead for assessing cell viability and phalloidin for assessing cell morphology. Use 400 µl as the volumes for all the solutions in the staining protocols and add all the solutions in the upper channel of the chips.
- For Live/Dead staining, aspirate the medium from the chips and wash with DPBS for 5 min then add the staining solution of Calcein-AM and ethidium homodimer-1 for 30-40 min. Following staining, remove the solution and wash with DPBS. Store in DPBS in the channel and image with fluorescent microscope.
- For phalloidin staining, aspirate the medium from the chips and wash with DPBS for 5 min and then fix with 4% PFA for 30 min. Wash twice with DPBS and permeabilize with 0.1% Triton X-100 in DPBS for 10 min. Wash three times with DPBS and block with 1% BSA in DPBS for 2 h. Stain with solution containing phalloidin-TRITC and DAPI for 2 h, then wash four times with DPBS for 30 min-1 h. Store in DPBS in the channel and image with fluorescent microscope.
RESULTS
BMSCs were successfully embedded in GrowDex-T, fibrin and collagen I at a density of 1.5 x 106 cells/ml and loaded into the bottom chamber of the BE-Doubleflow microfluidic chip (Figure 1). Cell culture media was passed through the top chamber and all hydrogel cultures withstood a fluidic flow rate of 12 µl/min applied for 7 days of culture. Static controls, where no flow was applied, were also successfully established.
The cells remained viable and were clearly detectable when observing the cells using fluorescent and brightfield microscopes. Clear fluorescent images of cells embedded in all three hydrogels were collected following Live/Dead and phalloidin staining (Figure 4).
GrowDex and fibrin were more optically clear when compared with collagen I. The 3D structure of the cells was more challenging to distinguish when cultured in collagen I, especially following fluidic flow (Figure 4). Although minor shrinkage or hydrogel rearrangement by the cells was seen with the GrowDex®-T cultures, the hydrogel remained intact through the end of the culture period. With collagen I, hydrogel shrinkage was more drastic, whereas fibrin showed no signs of shrinkage or hydrogel rearrangement.
Following phalloidin staining (Figure 4), no major differences in cell morphology were seen when cultured in any of the three hydrogels. It was seen that the cells displayed an elongated morphology and were in close contact with each other in the 3D environment. Due to the collagen I hydrogel shrinkage, cells were seen to be more compressed and closer together, with some alignment, however this was not quantified.
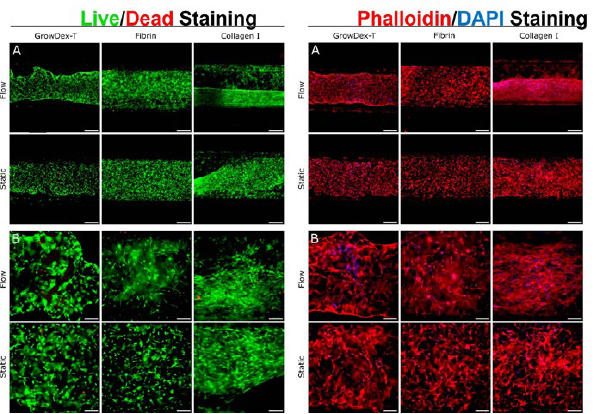
Figure 4.
Live/dead staining (left panels) and phalloidin/DAPI staining (right panels) of BMSCs embedded in GrowDex-T, fibrin, and collagen I when cultured for 7 days under active flow (Flow) or 8 days without flow (Static). Live/dead staining shows viable cells stained green with Calcein AM, and dead cells stained red with ethidium homodimer-1. Phalloidin/ DAPI staining shows actin cytoskeleton stained red, and cell nuclei blue. Scale bar represents 500 µM in panel A and 200 µM in panel B.
CONCLUSIONS
In conclusion, BMSCs were successfully embedded in GrowDex-T and fibrin and collagen I and cultured under active flow for 7 days. Collagen I underwent major shrinkage and reshaping during the culture period, especially when cultured under flow conditions. Some reshaping and shrinkage of GrowDex-T was observed, most likely due to cell reorganization and ECM production, however further experiments will be needed to confirm this. With the addition of aprotinin, no shrinkage or matrix degradation of the fibrin hydrogel was observed.
Live/Dead viability analysis showed that BMSCs were viable in all three hydrogels throughout the culture period, in both static and under active flow conditions. Longer experiments could be conducted using the setup established here.
GrowDex-T shows great potential as an animal-free option for culturing BMSCs under flow conditions. Additionally, with the combination and co-culture of other cell types, this culture method allows for the establishment of more complex, in vivo-like tissue structures for OOC studies, recreating the required complexity of the cell’s microenvironment.
REFERENCES
- Kapałczyńska, M., et al. (2018). "2D and 3D cell cultures - a comparison of different types of cancer cell cultures." Archives of medical science : AMS 14(4): p. 910-919.
- van der Meer, A.D. and van den Berg, A. (2012). "Organs-on-chips: breaking the in vitro impasse." Integr Biol (Camb) 4(5): p. 461-70.
- Langhans, S.A. (2018). "Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning." Frontiers in Pharmacology 9.
- Hughes, C.S., et al. (2010). "Matrigel: A complex protein mixture required for optimal growth of cell culture." PROTEOMICS 10(9): p. 1886-1890.
- Van Norman, G.A. (2019). "Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Is it Time to Rethink Our Current Approach?" JACC: Basic to Translational Science 4(7): p. 845-854.
- UPM-Biomedicals. (2021). "GrowDex® Hydrogel Range." UPM Biomedicals Brochure. Retrieved 17/05/2022, from https://www.upmbiomedicals.com/siteassets/documents/growdex-hydrogels-product-brochure-2021.pdf.
- Kalke, E., et al. "GrowDex®-T in a commercial organ-on-chip under active flow." GrowDex-T Application Note 11.
- Chou, D.B., et al. (2020). "On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology." Nature Biomedical Engineering 4(4): p. 394-406.
- BEOnChip. (2021). "Be-Doubleflow: User Guide." BEOnChip User Guide. Retrieved 17/05/2022, from https://beonchip.com/wp-content/uploads/2021/09/BDF-V7-estandar.pdf